Our Research
The Ohainle Lab uses functional genomics and molecular virology as tools to understand how key molecular interactions between host and viral proteins mediate outcomes of infection.
We study this in the context of innovation in both host and viral genomes across evolutionary time. Our work focuses on the important human pathogen HIV and related retroviruses as a model to understand host/pathogen biology more broadly.
HIV Capsid-Targeting Restriction Factors
After fusing with the target cell membrane the core of HIV must transit through the cytoplasm to the nucleus and through the nuclear pore in order to integrate a DNA copy of its genome into host cell genome.
A single HIV protein, the capsid, must engage with host cell factors to faciltate infection while also evading host antiviral proteins that have evolved to target invading viruses. We are interested in understanding the mechanisms through which these antiviral proteins inhibit viral infection and how host and virus adapt in this arms race.
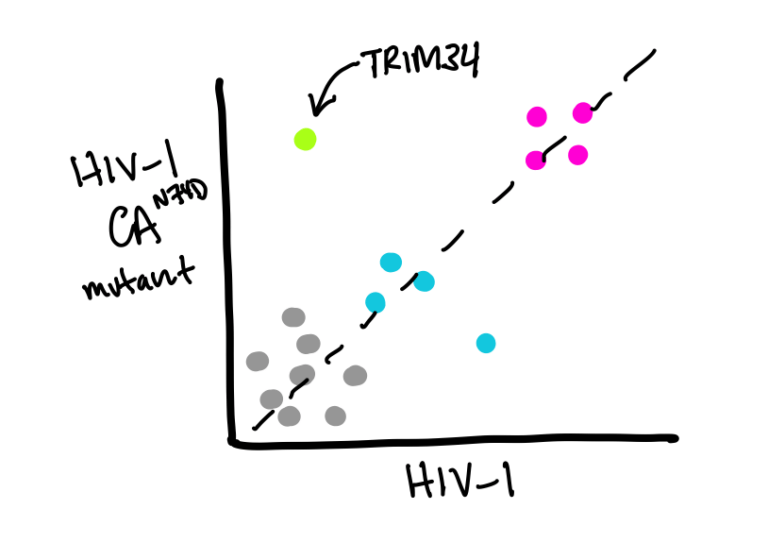
Evolution and Mechanism of TRIM restriction
We recently identified a new component (TRIM34) of a potent antiviral restriction mechanism that acts to recognize and inhibit the HIV core in the cytoplasm.
We are interested in understanding the mechanisms by which TRIM34 works with TRIM5 to inhibit retroviruses, when and where HIV is sensitive to this block and how TRIM34’s evolution in primates has contributed to blocks to cross-species transmission of retroviruses
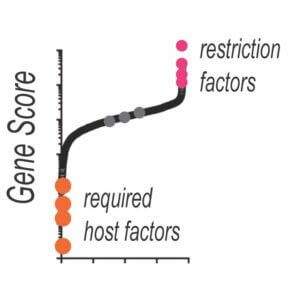
Comprehensive Restriction Factor Discovery
Through new directions in HIV-CRISPR screening we aim to comprehensively define antiviral proteins that inhibit replication of HIV and related retroviruses in human cells.
We are approaching this with modified CRISPR technologies, application of HIV-CRISPR screening to new cells types and conditions as well as through a more broad analysis of viral strains and mutants that we believe will allow for a more complete understanding of host cell restriction mechanisms.
Functional Genomics Tools in Virology
Our lab is interested in developing “massively parallel” approaches as tools for identifying key molecular interactions between host and virus proteins.
In addition to studying contemporary and retrospective host and virus protein interactions we are interested in further developing tools to define the molecular rules that determine outcomes of interactions at these interfaces.
We are pursuing new directions in virus CRISPR screening methods as well as developing other functional genomics approaches to study host/virus biology.
Other Directions
Interested in joining us? Contact Molly to discuss further project ideas.


